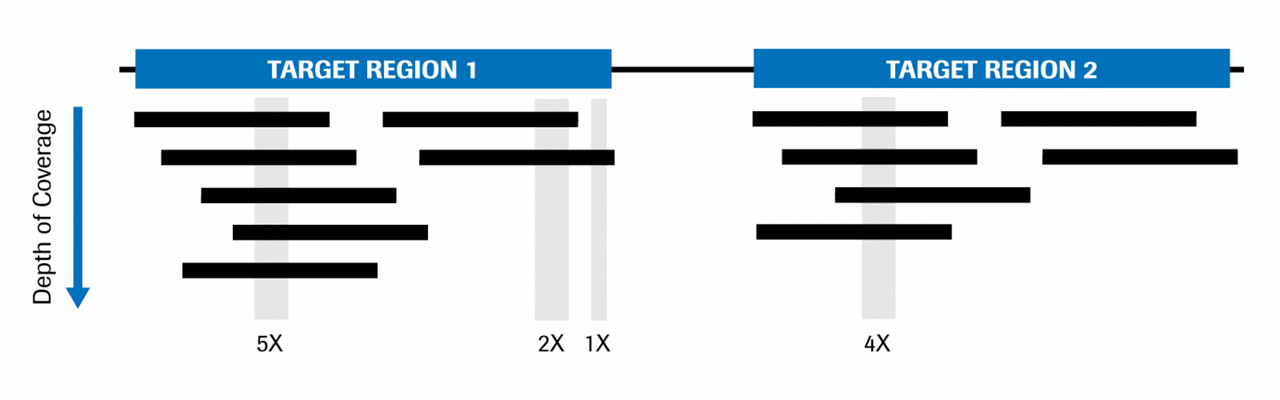
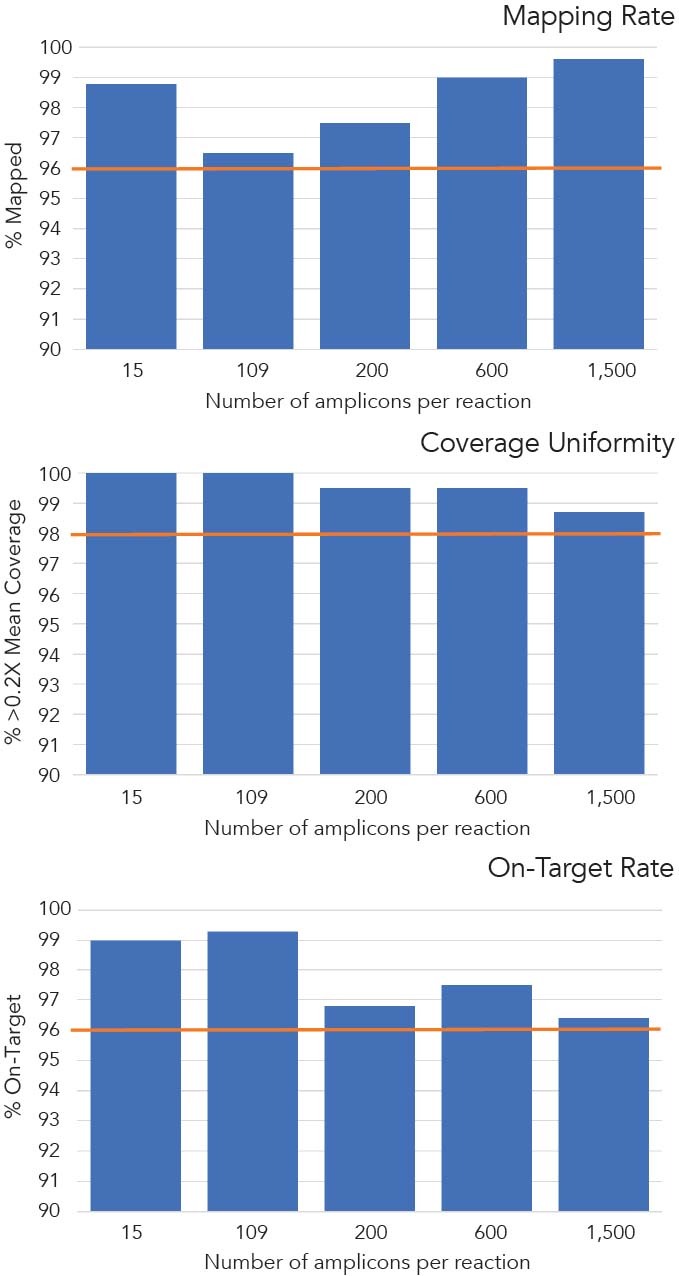
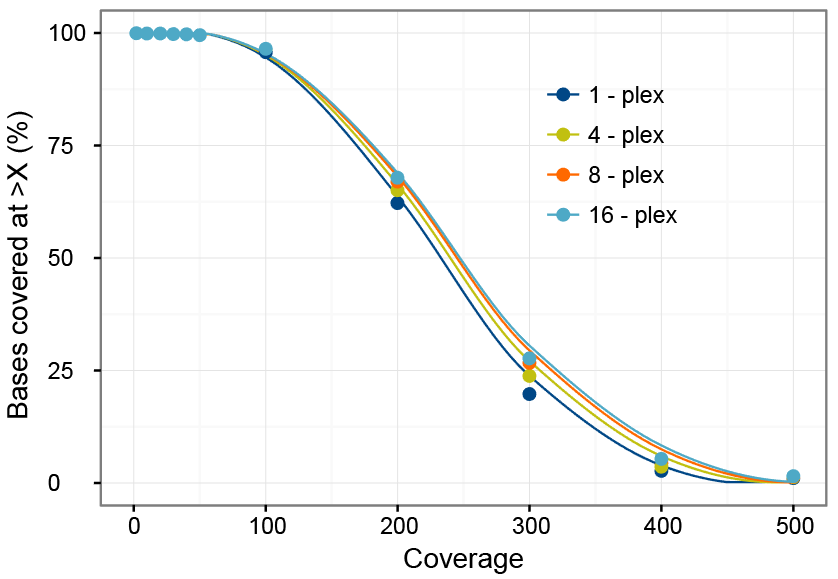
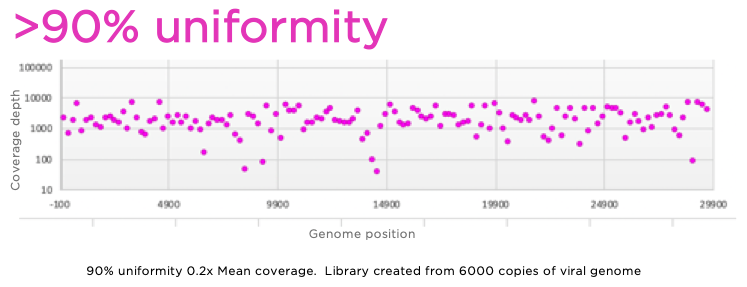
Method for improving sequence coverage uniformity of targeted genomic intervals amplified by LR-PCR using Illumina GA sequencing-by-synthesis technology | BioTechniques

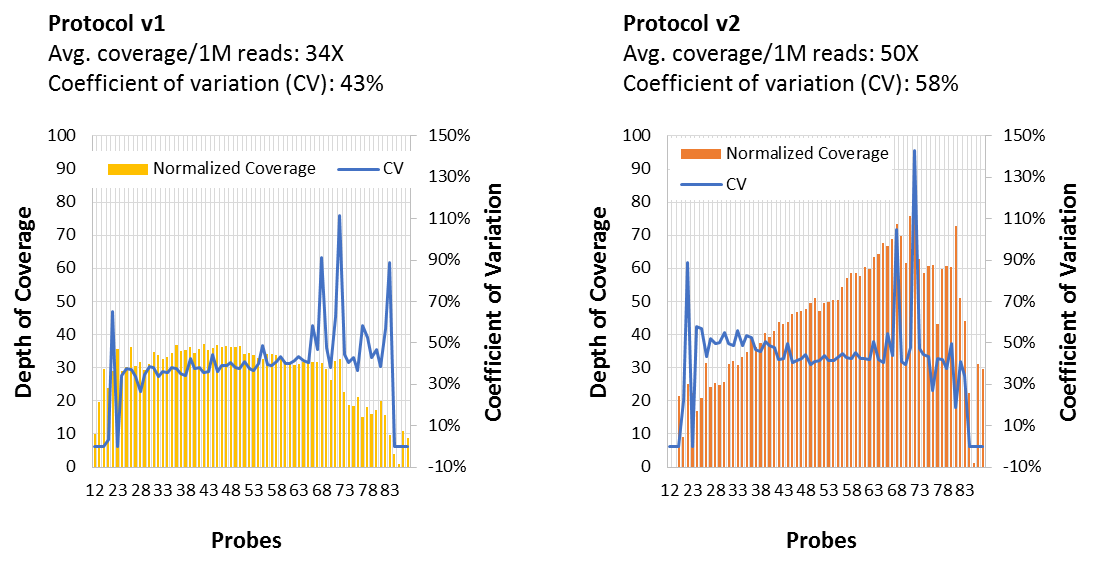
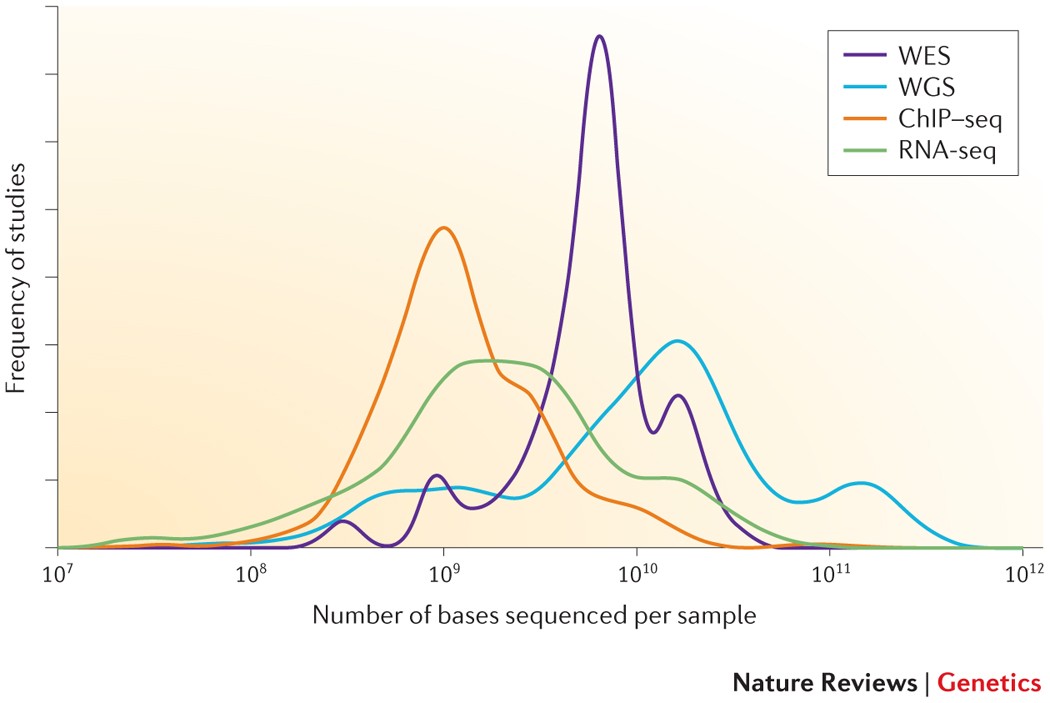
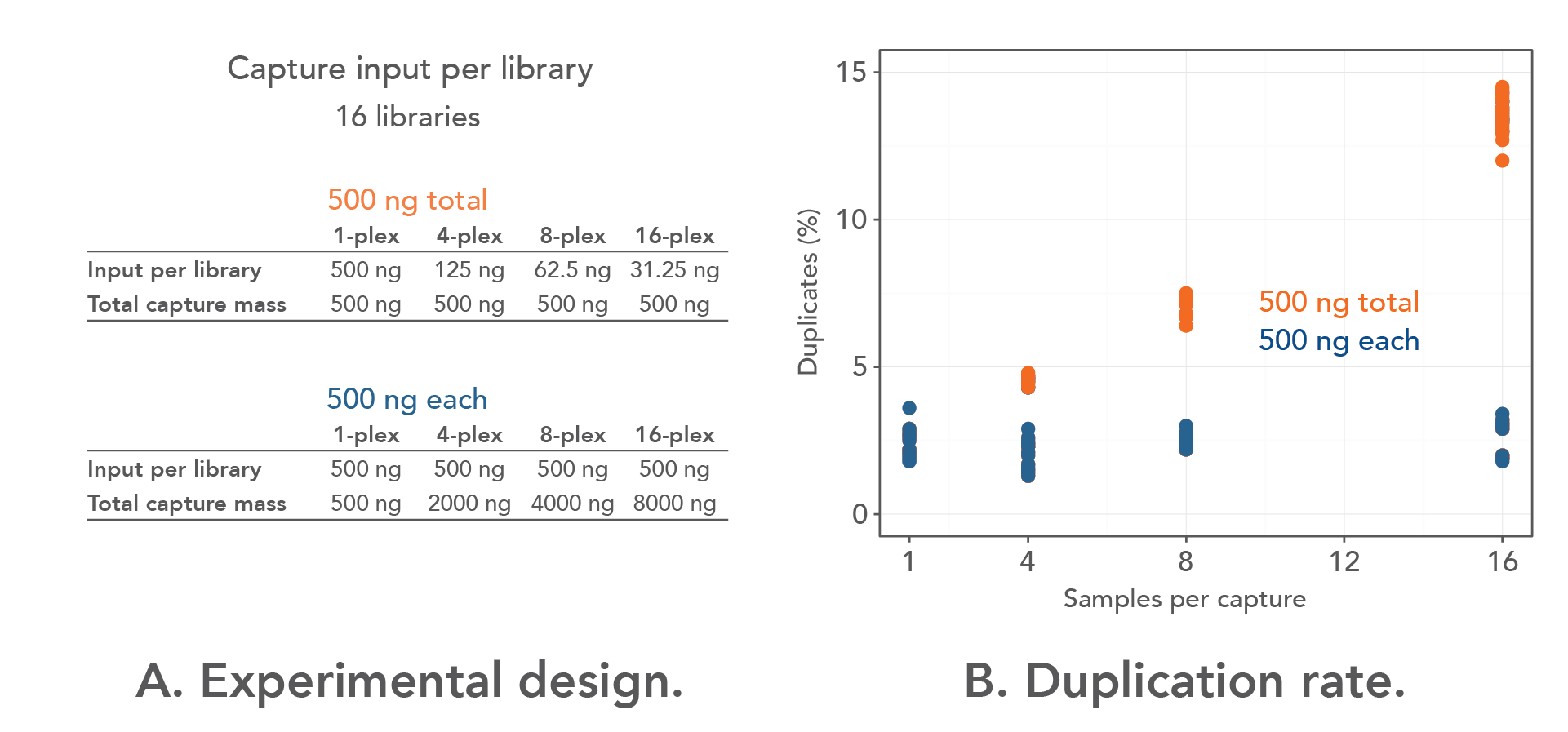
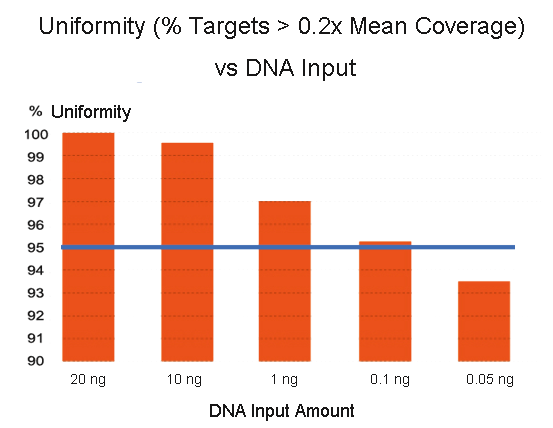
The Importance of Coverage Uniformity Over On-target Rate for Efficient Targeted NGS | Whitepaper | Technology Networks

Method for improving sequence coverage uniformity of targeted genomic intervals amplified by LR-PCR using Illumina GA sequencing-by-synthesis technology. | Semantic Scholar

Coverage uniformity of WGS libraries. a Represented is the cumulative... | Download Scientific Diagram

A Method to Evaluate the Quality of Clinical Gene-Panel Sequencing Data for Single-Nucleotide Variant Detection - ScienceDirect

Genes | Free Full-Text | Technological Advances: CEBPA and FLT3 Internal Tandem Duplication Mutations Can be Reliably Detected by Next Generation Sequencing
Amplicon-by-amplicon Read Coverage, Uniformity and Variant Detection in... | Download Scientific Diagram

A benchmarking study of SARS-CoV-2 whole-genome sequencing protocols using COVID-19 patient samples | bioRxiv